Liangliang Sun

Research
High-resolution, Ultrasensitive, and Native Proteomics
(Research Description PDF)
Proteomics aims to comprehensively identify and quantify proteins in a biological system, including protein expression, localization, interaction, posttranslational modifications (PTMs) and turn over. It routinely employs reversed-phase liquid chromatography (RPLC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/ MS) for protein identification. Capillary zone electrophoresis (CZE)-ESI-MS/MS has also attracted great attentions for proteomics due to its advantageous features. First, CZEMS and RPLC-MS can produce complementary identifications and the combination of these two techniques can improve proteomic scale, and especially enhance proteoform identifications. Second, CZE can produce better intact protein separation than RPLC, benefiting top-down proteomics. Third, CZE-MS can yield higher sensitivity than RPLC-MS for detection of peptides and intact proteins. Fourth, CZE can separate proteins under native conditions. CZE-MS/MS will be an invaluable tool for native proteomics that aims to approach proteomescale characterization of endogenous protein complexes in cells.
Our research focuses on development of novel analytical methodologies based on CZE-MS/ MS for high-resolution, ultrasensitive and native proteomics, and applications of the new methodologies for answering important questions in biology.
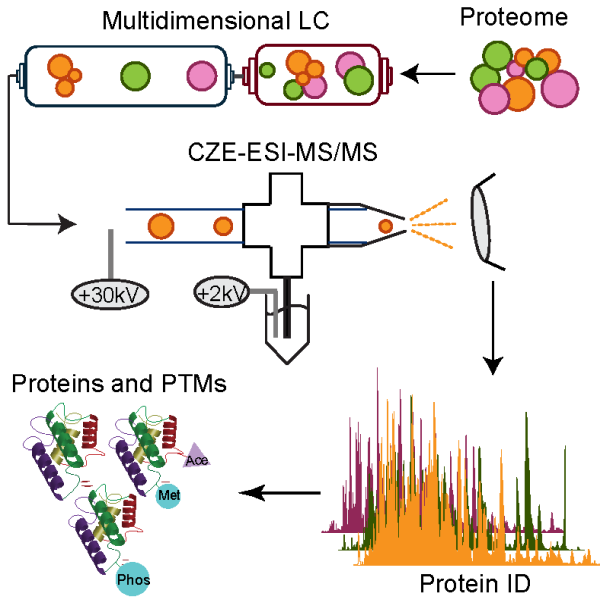
(I) Couple multi-dimensional LC to CZE-MS/MS for high-resolution and ultrasensitive proteomics. We employ orthogonal separation techniques to improve the separation of peptides and intact proteins in complex proteomes, boosting the proteome coverage from proteomics. We integrate microscale RPLC (µRPLC) with CZE-MS/MS to improve the sensitivity of proteomics, enabling deep proteomics of mass-limited samples. We collaborate with developmental biologists to apply our techniques for understanding important questions in vertebrate early embryogenesis using zebrafish as a model system. We are particularly interested in two important questions. First, how do proteins and/or their PTMs accurately control the zygotic genome activation at mid-blastula transition? Second, when and how do interblastomere differences arise during early cellular differentiation? We believe quantitative proteomics of zebrafish embryos and blastomeres across multiple developmental stages will provide valuable insight into those questions.
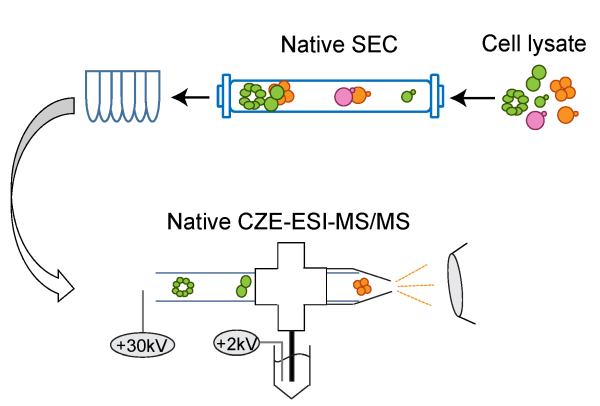
(II) Develop analytical methods for native proteomics. We couple size exclusion chromatography (SEC) to CZE-MS/MS for highresolution separation of complex proteomes under native conditions. The SEC-CZE-MS/ MS will enable large-scale identification and relative quantification of protein complexes directly from complex proteome samples and in discovery mode. We are particularly interested in characterization of protein-metal complexes in cells.
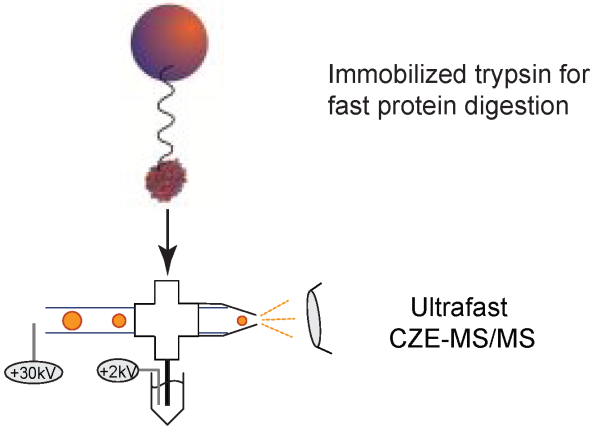
(III) Couple magnetic beads-based immobilized trypsin to fast CZE-MS/MS for high-throughput proteomics. The state-of-the-art proteomics platforms require at least 12 hours for sample preparation, RPLC-MS/MS, and data analysis. The relatively low throughput impedes application of proteomics for daily and system-wide clinical diagnostics. Our goal is to improve the throughput of proteomics by over one order of magnitude, approaching halfan- hour proteomics. We believe the technique will facilitate daily and system-wide clinical diagnostics
Contact / Webpage
Area(s) of Interest
Analytical (An)
Biological (Bi)
Material (Ma)
Selected Publications
Single-Shot Top-Down Proteomics with Capillary Zone Electrophoresis- Electrospray Ionization-Tandem Mass Spectrometry for Identification of Nearly 600 Escherichia coli Proteoforms, Lubeckyj, RA; McCool, EN; Shen, X; Kou, Q; Liu, X; Sun, L., Anal. Chem. 2017, 89, 12059-12067.
Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography-Capillary Zone Electrophoresis-Tandem Mass Spectrometry, Shen, X., Kou, Q., Guo, R., Yang, Z., Chen, D., Liu, X, Hong, H., Sun, L., Anal Chem. 2018, i, 10095-10099.
Deep Top-Down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia coli Proteome, McCool, EN; Lubeckyj, RA; Shen, X; Chen, D; Kou, Q; Liu, X; Sun, L; Anal. Chem. 2018, 90, 5529-5533.
Microscale Reversed-Phase Liquid Chromatography/Capillary Zone Elect rophore s i s -Tandem Mass Spectrometry for Deep and Highly Sensitive Bottom-Up Proteomics: Identification of 7500 Proteins with Five Micrograms of an MCF7 Proteome Digest, Yang, Z, Shen, X., Chen, D., Sun L., Anal Chem. 2018, 90(17), 10479-10486.
Capillary zone electrophoresis-tandem mass spectrometry for large-scale phosphoproteomics with the production of over 11000 phosphopeptides from the colon carcinoma HCT116 cell line, Chen, D., Ludwig, K., Krokhin, O.V., Spicer, V., Yang, Z., Shen, X., Hummon, A.B., Sun, L., Anal Chem. 2019, 91(3), 2201-2208.
CV
B.S., 2005, Dalian University of Technology;
Ph.D., 2011, Dalian Institute of Chemical Physics, Chinese Academy of Sciences
Postdoctoral Fellow (2011-12) and Research Assistant Professor (2013-16), Univ. of Notre Dame
Awards
| Year | Award | Organization |
|---|---|---|
| 2021 | Thermo Fisher Scientific Early Career Award | |
| 2019 | Emerging Investigator | Royal Society of Chemistry journal Analytical Methods |
| 2019 | NSF Early CAREER Award | |
| 2019 | Emerging Investigator | Journal of The American Society for Mass Spectrometry (JASMS) |