Heedeok Hong

Research
Biophysical Chemistry of Membrane Protein Folding and Degradation
(Research Description PDF)
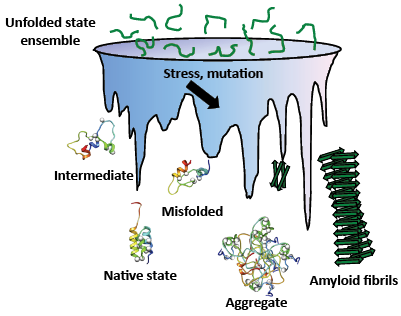
Protein folding is an amazing molecular process that occurs by sorting out an astronomical number of possible conformations down the free energy landscape. In a crowded cellular environment, however, environmental stresses or mutations can mislead polypeptide chains to misfolded or catastrophic aggregate states (Fig. 1). Therefore, these unnecessary proteins have to be selectively cleared from cells for quality control and regulatory purposes. For the past decades, there have been remarkable advances in understanding these phenomena and related diseases. However, efforts have been largely limited to water-soluble proteins excluding the other major class of proteins that reside in cell membranes.
Our research focuses on a fundamental biological question, how membrane proteins are made and destroyed in cells (Fig. 2). Membrane proteins comprise approximately 30% of all proteins encoded in genes and carry out numerous critical cellular functions. Approximately 60% of all drug developments target membrane proteins. The folding problem of membrane proteins is directly connected to human health. Indeed, accumulation of misfolded or misprocessed membrane proteins causes serious diseases such as Alzheimer’s disease, cystic fibrosis, and cancer. To answer our cardinal question, we investigate two conceptually connected biological processes by multi-disciplinary approaches including biochemical, biophysical, and chemical methods.
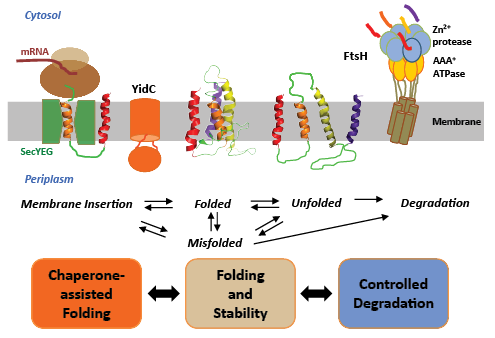
Chaperone-assisted membrane protein folding: YidC/Oxa1/Alb3 is a membrane protein family that plays a critical role in folding and assembly of membrane proteins in the inner membranes of bacteria, mitochondria, and chloroplasts. In E. coli, YidC forms a membrane insertion pore independent of SecYEG complex, major protein translocation machinery. YidC also has a chaperone activity: it facilitates the folding of a variety of SecYEG-dependent proteins. To understand how YidC acts as chaperone, we will tackle three specific problems:
- What are the driving forces in YidC-substrate interaction?
- What mechanism does YidC use to facilitate folding of membrane proteins?
- How are the structure and dynamics of YidC related to the function?
Controlled degradation of membrane proteins: Rapid protein degradation is a crucial cellular process that enables the clearance of misfolded proteins and regulatory proteins that are no longer needed. In all cells, this process is mediated by AAA+-protease superfamily. FtsH is the only membrane-localized AAA+-protease, which degrades both membrane and cytosolic proteins. To understand the principles of the quality control mechanism of membrane proteins, we focus on three specific questions using FtsH from E. coli as model.
- What sequence or structural features of substrates are subject to degradation?
- What is the role of the FtsH transmembrane domain in recognition and translocation of substrates?
- How is the proteolytic activity modulated by other membrane-bound cofactors?
Graduate students will gain a training opportunity in DNA manipulation, expression and purification of membrane proteins, biophysics of lipid bilayers, protein labeling, and various biophysical tools such as fluorescence, EPR, and X-ray crystallography.
Contact / Webpage
Area(s) of Interest
Biological (Bi)
Physical (Ph)
Selected Publications
The lipid bilayers strengthens the cooperative network of membrane proteins, MuhammedNazaar, S., Yao, J., Necelis, M.R., Park, Y.C., Shen, Z., Bridges, M.D., Guo, R., Swope, N., Rhee, M.S., Kim, M., Kim, K.H., Hubbell, W.L., Fleming, K.G., Columbus, L., Kang, S.-g., and Hong, H., Science Advances 2025, 11, eadv9568.
Lipid bilayer induces contraction of the denatured state ensemble of a helical-bundle membrane protein, Gaffney, K.A., Guo, R., Bridges, M.D., Muhammednazaar, S., Chen, D., Kim, M., Yang, Z., Schilmiller, A.L., Faruk, N.F., Peng, X., Jones, A.D., Kim, K.H., Sun, L., Hubbell, W.L., Sosnick, T.R., and Hong, H., Proc. Natl. Acad. Sci. USA 2022, 119, e2109169119.
Untangling the complexity of membrane protein folding, Hong H, Choi HK, and Yoon T.-Y, Curr. Opin. Struct. Biol. 2022, 72, 237-247.
Structural cavities are critical to balancing stability and function of a membrane-integral enzyme, Guo, R., Cang, Z., Yao, J., Kim, M., Deans, E., Wei, G., Kang, S.-g., and Hong, H., Proc. Natl. Acad. Sci. USA 2020, 117, 22146-22156.
Proteolysis mediated by the membrane-integrated ATP-dependent protease FtsH has a unique nonlinear dependence on ATP hydrolysis rates, Yang, Y., Gunasekara, M., Muhammednazaar, S., Li, Z. and Hong, H. Protein Sci. 2019, 28,1262-1275.
The rhomboid protease GlpG has weak interaction energies in its active site hydrogen bond network, Gaffney, K. and Hong, H., J. Gen. Physiol. 2018, 151, 282-291. (Highlighted in JGP research news.)
Folding-degradation relationship of a membrane protein mediated by the universally conserved ATP-dependent protease FtsH, Yang, Y., Guo, R., Gaffney, K., Kim, M., Muhammednazaar, S., Wang, B., Wei, T., Liang, J., and Hong, H., J. Am. Chem. Soc. 2018, 140, 13, 4656-4665. (Selected as a JACS spotlight article.)
Steric trapping reveals a cooperative network in the intramembrane protease GlpG, Guo, R., Gaffney, K.A., Kim, M., Yang, Z., Sungsuwan, S., Huang, X., Hubbell, W.L. and Hong, H., Nature Chem. Biol. 2016, 12, 353-36
CV
B.S. 1996, M.S. 1998,Yonsei University, South Korea
Ph.D., 2006, Univ. of Virginia
Postdoctoral Fellow, 2006-2012, University of California, Los Angeles
The Leukemia and Lymphoma Society Postdoctoral Fellowship, 2008-2011