James H. Geiger

Research
Structural Biology Using X-Ray Crystallography
(Research Description PDF)
The major focus of our research is to both understand and engineer proteins and protein complexes at atomic resolution. This allows us to both think about and manipulate these large molecular systems chemically, with an emphasis on how the atomic resolution structural details of the systems give rise to their properties, activities and function.
Structure and Mechanism of Enzymes. We have determined the structures of all three of the enzymes in the starch biosynthetic pathway, ADP-glucose pyrophosphorylase, Branching enzyme and glycogen/starch synthase. Our structures of glycogen synthase and branching enzyme showed for the first time that several glycogen binding sites exist outside the enzyme’s active site. They are important for the enzyme’s activity and understanding their role is a focus of our future work. More recently we have determined a structure of rice Branching enzyme bound to maltododecaside, which unveiled the trajectory of the glucans that react. We now understand the determinants of substrate specificity for this, and essentially all the other branching enzymes. This allows us to start thinking about how to predict the reactivity of these enzymes from their sequence. In collaboration with the Walker lab, we have also investigated the structure, mechanism and specificity of some of the enzymes involved in Taxol biosynthesis, including phenylalanine aminomutase (PAM) and benzoic acid CoA ligase. The structures of 2 PAMs have been determined, and a variety of benzoic acid CoA ligase substrate structures have been determined for use in rationally extending the substrate specificity of these enzymes.
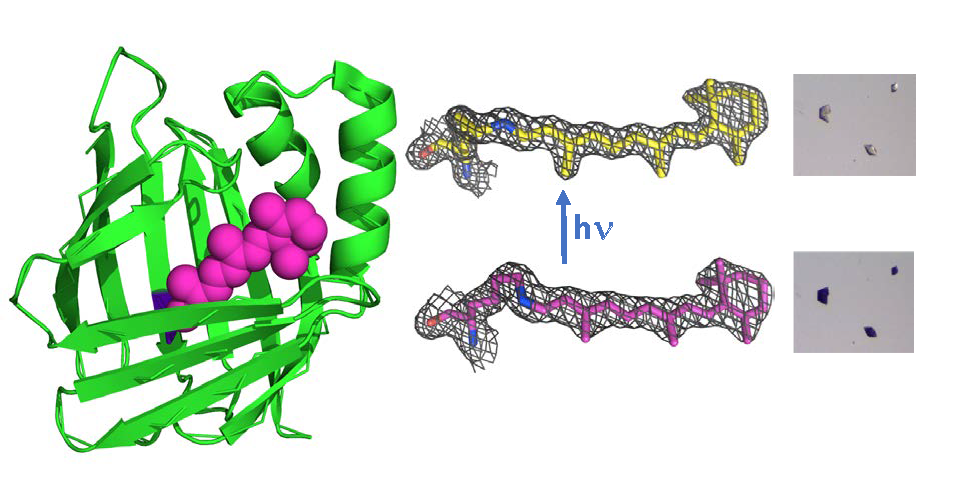
Visualizing rhodopsin mimics at atomic resolution. In collaboration with the Borhan lab, we redesigned small cytosolic proteins (cellular retinoic acid binding protein II (hCRABPII) and cellular retinol binding protein II (hCRBPII) to be rhodopsin mimics that form a protonated Schiff base with the retinal chromophore. Further, we constructed a spectrum of protein pigments that bind the same chromophore retinal, but alter the absorbance of this chromophore over 219 nm. We have also developed new fluorescent proteins that can be used as fluorescent protein fusion tags, extending the range of fluorescent proteins and adding pH sensing to their repertoire. More recently we have turned our attention to understanding the photoisomerization process. To this end we have engineered retinal-bound protein variants that specifically photo-isomerize only one of the five double bonds in the chromophore. So far we have variants that specifically isomerize either the imine bond or the 13-bond, the latter of which is the very same bond isomerized in all bacterial rhodopsins. Further, we observe these isomerizations in single crystals, at atomic resolution. We are now pursuing timeresolved crystallographic experiments on these species to visualize the intermediates in the photo-isomerization process. There are very few systems that lend themselves to the difficult techniques of time resolved crystallography and our model systems are ideal candidates. The ability to design from scratch protein/chromophore systems that work so spectacularly open the door to creating completely novel photo-active proteins for a wide variety of potential applications.
Engineering domain swapped dimers and the creation of new allosteric proteins. We have recently discovered that variants of hCRBPII readily and irreversibly fold into domain swapped dimers (DS dimers). A DS dimer forms when identical elements of structure swap places in two monomers to form a dimeric structure. After characterizing some of the determinants of DS dimer formation, so that we can control its formation, we discovered that ligand binding induces a dramatic conformational change in the DS dimer. A variety of ligands illicit the conformational change, including retinal, fatty acids and some fluorophores. We are currently using the DS dimer to create new allosteric proteins that can be controlled by redox state, ligand or metal binding depending on our design. Previous efforts at allosteric protein design use naturally occurring allosteric proteins to control other processes. This is one of the few cases where an allosteric protein is engineered “from scratch.”
Contact / Webpage
Area(s) of Interest
Physical (Ph)
Biological (Bi)
Selected Publications
Photoisomerizing Rhodopsin Mimic Observed at Atomic Resolution, Nosrati, M., Berbasova, T., Vasileiou, C., et al., J. Am. Chem. Soc. 2016, 138, 8802-8808.
Crystal structures of Escherichia coli branching enzyme in complex with cyclodextrins, Feng, L., Fawaz, R., Hovde, S., et al., Acta Crystallographica Section D-Structural Biology 2016, 72, 641-647.
Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates, Assar, Z., Nossoni, Z., Wang, W., et al., Structure 2016, 24, 1590-1598.
Stereochemistry and Mechanism of a Microbial Phenylalanine Aminomutase, Ratnayake, N. D.; Wanninayake, U.; Geiger, J. H.; Walker, K. D., J. Am. Chem. Soc. 2011, 133 (22), 8531-8533.
"Turn-On" Protein Fluorescence: In Situ Formation of Cyanine Dyes, Yapici, I., Lee, K. S. S., Berbasova, T., et al., J. Am. Chem. Soc. 2015, 137, 1073-1080.
Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity, Thornburg, C. K., Wortas-Strom, S., Nosrati, M., et al., Biochemistry 2015, 54, 6230-6242.
Tuning the Electronic Absorption of Protein-Embedded All-trans-Retinal, Wang, W., Nossoni, Z., Berbasova, T., et al., Science 2012, 338, 1340-1343.
Insights into the Mechanistic Pathway of the Pantoea agglomerans Phenylalanine Aminomutase, Strom, S., Wanninayake, U., Ratnayake, N. D., et al., Angew. Chem.-Int. Ed. 2012, 51, 2898-2902.
Light-Activated Reversible Imine Isomerization: Towards a Photochromic Protein Switch, Berbasova, T., Santos, E. M., Nosrati, M., et al., Chembiochem 2016, 17, 407-414.
CV
B.S., 1984, New College of USF (Florida)
Ph.D., 1991, Princeton Univ.
American Cancer Society Postdoctoral Fellow 1994, Yale Univ.
Postdoctoral Fellow, 1996, Yale Univ.
Awards
| Year | Award | Organization |
|---|---|---|
| 1994 - 1995 | Postdoctoral Fellow | American Cancer Society |
| 1991 | Ph.D. | Princeton University |
| 1989 - 1996 | Postdoctoral Fellowship | Yale University |
| 1984 | Bachelor of Science | New College of University of South Florida |
| 1980 - 1984 | Marie Selby Full Tuition Scholarship | New College of University of South Florida |