Mary C. Andorfer

Research
Enzyme Engineering, Organic Chemistry, Structural Biology
(Research Description PDF)
The Andorfer lab seeks to develop sustainable biocatalysts for producing value-added chemicals from inexpensive and abundant feedstocks, namely CO2 and hydrocarbons. We will target enzymes that allow anaerobic microbes to activate components of crude oil (e.g. toluene, n-alkanes, and benzene) and subsequently use them as carbon sources for metabolism. Because anaerobic, crude-oil-polluted environments are recalcitrant to typical remediation methods, these enzymes could be useful for bioremediation efforts in anaerobic environments as well. The challenging task of anaerobic hydrocarbon activation is enabled by a range of enzyme cofactors, including both inorganic cofactors (e.g. 4Fe4S clusters) and/or organic radical cofactors (e.g. amino acid radicals). We aim to not only engineer these enzymes for selective catalysis, but also to elucidate molecular-level mechanistic understanding.
We will take two major approaches to study these crude oil degrading enzymes:
- Directed evolution of selective C–H functionalization biocatalysts for synthetically-useful transformations
- Structural and mechanistic characterization of putative hydrocarbon-degrading enzymes to understand basic principles of how they work
In addition to more traditional organic and molecular biology techniques, members of the group will use single particle cryo-electron microscopy (cryo-EM), EPR spectroscopy, as well as anaerobic enzyme purifications and biochemical assays.
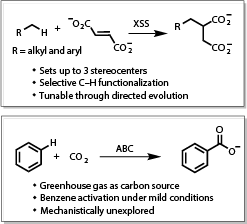
Directed Evolution of Anaerobic Enzymes - Directed evolution methodology has been used for decades to repurpose enzymes for synthetically useful organic transformations; however, it has not been used for anaerobic hydrocarbon activating enzymes due to the complexity of these systems. We will create tools to engineer these enzymes to alter functionality and develop them as synthetically useful catalysts. Some of the reactions we will be exploring will include olefin hydroalkylation and benzene carboxylation.

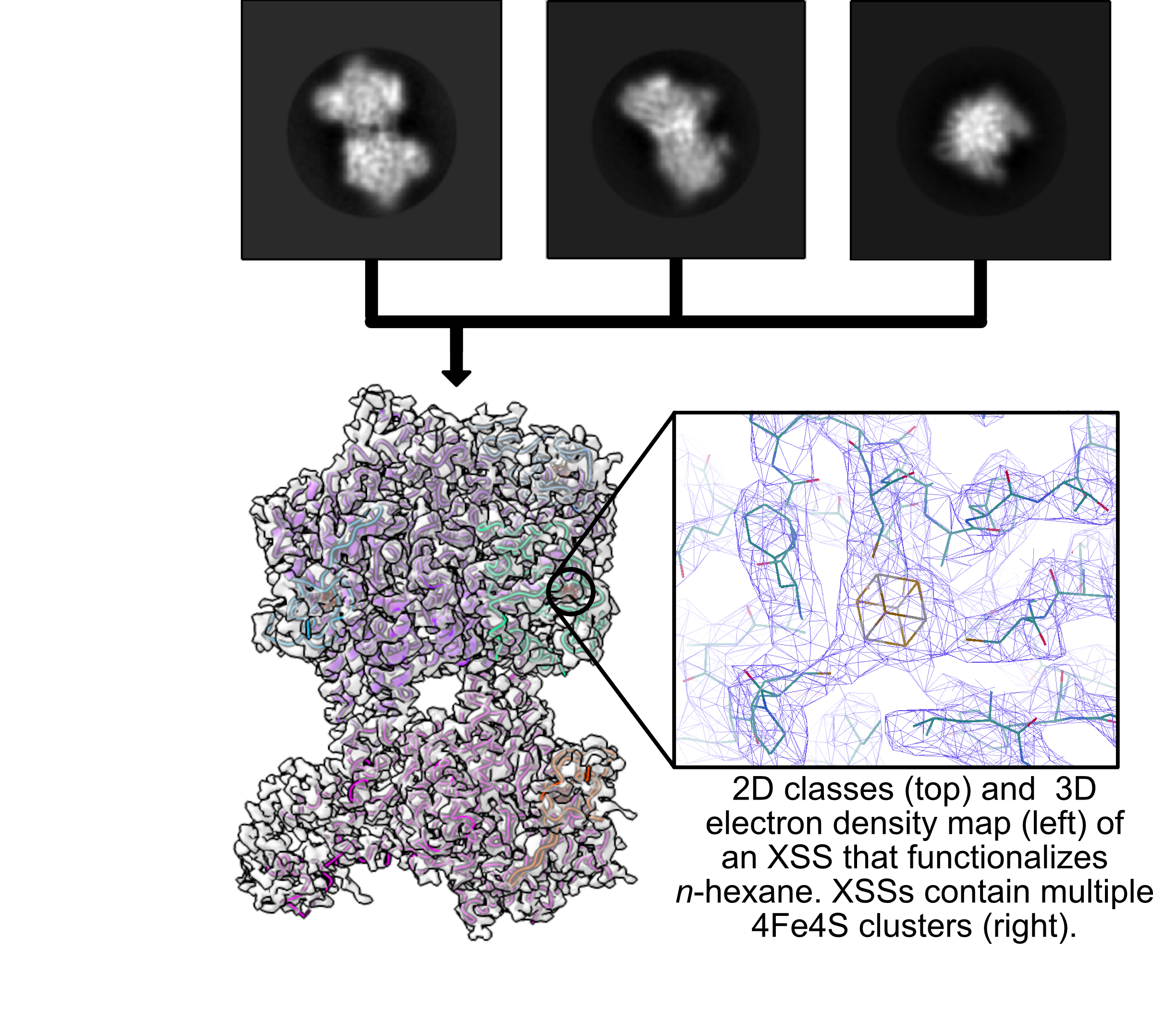
Single Particle Cryo-EM - We will use MSU’s Talos Arctica electron microscope to collect images to visualize single protein molecules. Single particles can be sorted into 2D classes and used to build 3D electron density maps for the protein of interest. Using these maps, we can build in molecular models to ultimately solve new protein structures. We will leverage the molecular-level mechanistic understanding gained through structure determination to inform protein engineering efforts.
Contact / Webpage
Area(s) of Interest
Organic (Or)
Biological (Bi)
Inorganic (In)
Selected Publications
Rescuing Activity of Oxygen-Damaged Pyruvate Formate-Lyase by a Spare Part Protein, Andorfer, M.C.; Backman, L.R.F.; Li, P.L.; Ulrich, E.C.; Drennan, C.L., J. Biol. Chem. 2021, 297, 101423.
Fixing Nature’s Carbon Inefficiencies, Andorfer, M.C.; Drennan, C.L., Joule 2021, 5, 765.
Molecular Basis for Catabolism of the Abundant Metabolite trans-4-hydroxyL-proline by a Microbial Glycyl Radical Enzyme, Backman, L.R.F.; Huang, Y.Y.; Andorfer, M.C.; Gold, B.; Raines, R.T.; Balskus, E.P.; Drennan, C.L., eLife 2020, 9, e51420.
Solution Structure and Biochemical Characterization of a Spare Part Protein that Restores Activity to an Oxygen-Damaged Glycyl Radical Enzyme, Bowman, S.E.J.; Backman, L.R.F.; Bjork, R.E.; Andorfer, M.C.; Yori, S.; Caruso, A.; Stultz, C.M.; Drennan, C.L., J. Biol. Inorg. Chem. 2019, 24, 817.
Understanding and Improving the Activity of Flavin Dependent Halogenases via Random and Targeted Mutagenesis, Andorfer, M.C.; Lewis, J.C., Annu. Rev. Biochem. 2018, 87, 159.
Understanding Flavin-Dependent Halogenase Reactivity via Substrate Activity Profiling, Andorfer, M.C.; Grob, J.E.; Hajdin, C.E.; Chael, J.; Siuti, P.; Lilly, J.; Tan, K.L.; Lewis, J.C., ACS Catal. 2017, 7, 1897.
Directed Evolution of RebH for CatalystControlled Halogenation of Indole C–H Bonds, Andorfer, M.C.; Park, H.J.; VergaraColl, J.; Lewis, J.C., Chem. Sci. 2016, 7, 3720.
Improving the Stability and Catalyst Lifetime of the Halogenase RebH By Directed Evolution, Poor, C.B.; Andorfer, M.C.; Lewis, J.C., ChemBioChem 2014, 15, 1286.
Regioselective Arene Halogenation using the FAD-Dependent Halogenase RebH, Payne, J.T.; Andorfer, M.C.; Lewis, J.C. Angew. Chem. Int. Ed. 2013, 52, 5271.
CV
B.S., Butler Univ.
Ph.D., Univ. of Chicago
Postdoctoral Fellow, Massachusetts Inst. of Technology