MSU Researchers bettered our understanding of colorectal cancer metastasis via comprehensive measurements of proteoforms in colorectal cancer cells
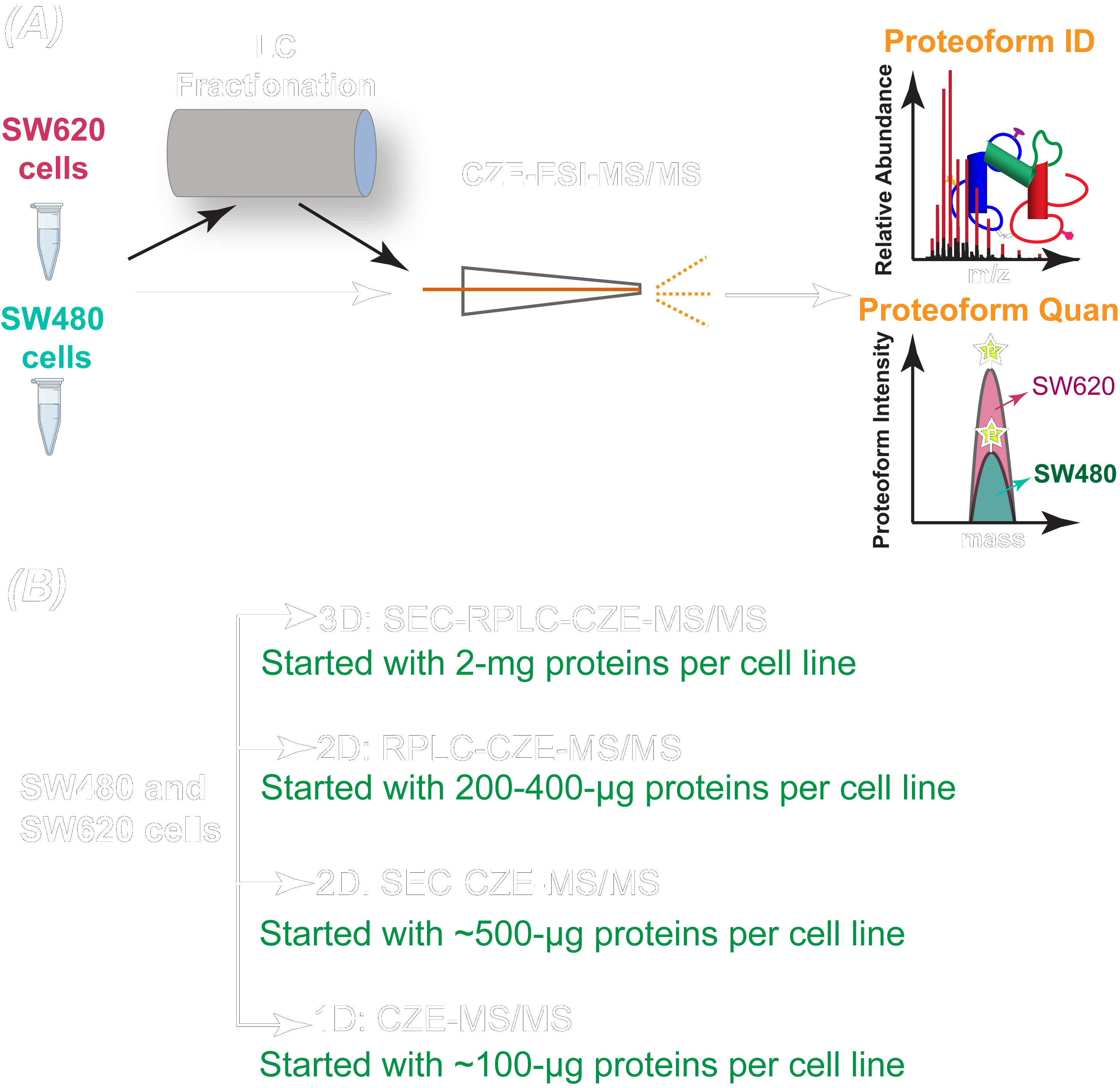
Studying cancer metastasis at the proteoform (all forms of protein molecules produced from the same gene) level will provide a more accurate understanding of the molecular mechanisms and discover novel protein biomarkers for cancer diagnosis and drug development. Genetic variation, alternative splicing, and post-translational modification work together to create a richer set of proteins, or proteoform, than strictly available from genetically encoded proteins.
Dr. Liangliang Sun’s group at MSU Chemistry recently reported in Science Advances the first deep top-down proteomics study of a pair of isogenic human nonmetastatic and metastatic colorectal cancer cell lines with the identification of over 23,000 proteoforms of more than 2,000 genes from the two cell lines. This represents a pivotal advance in mass spectrometry-based top-down proteomics toward the characterization of human proteome in a proteoform-specific manner.
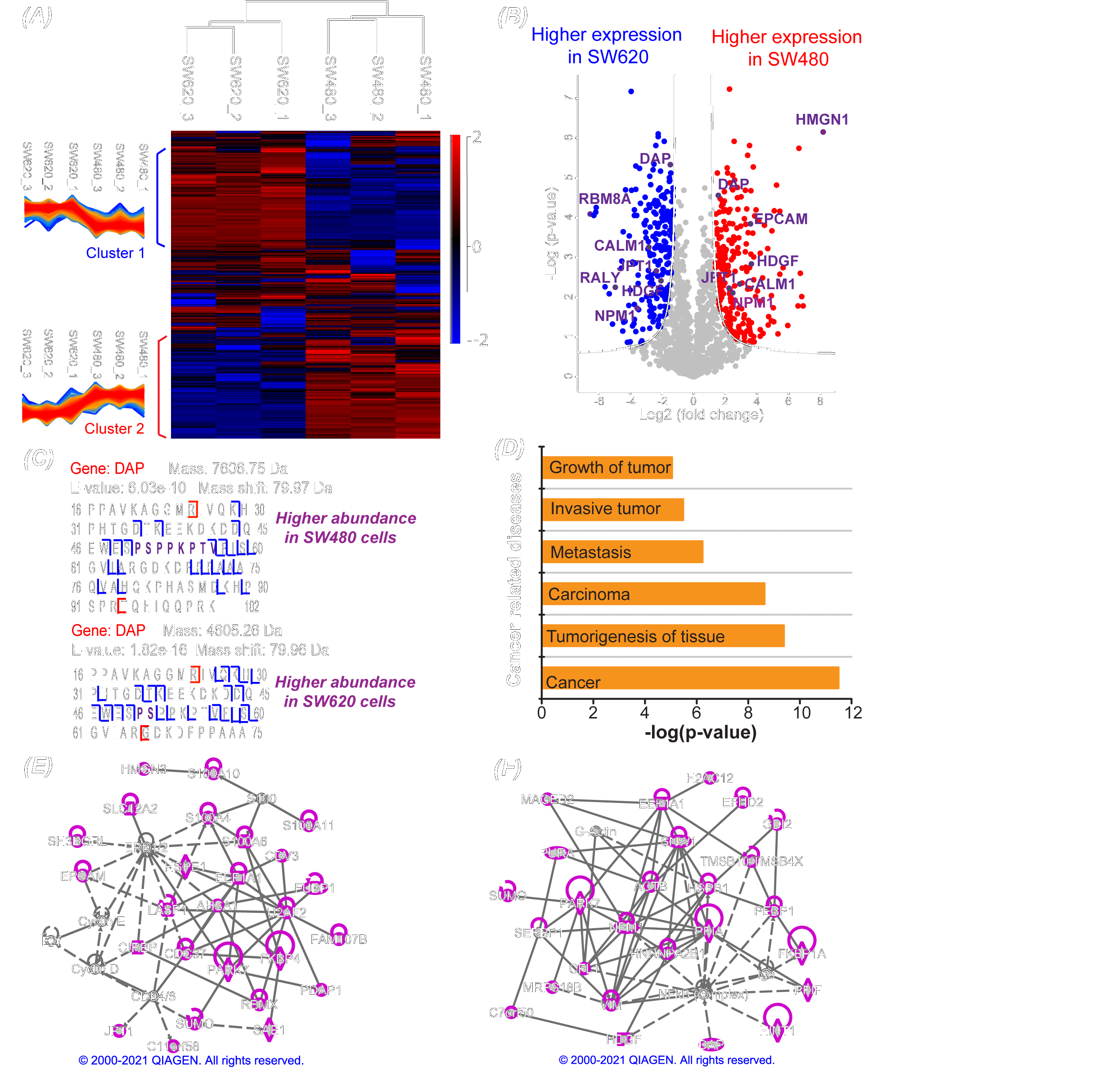
The study revealed the drastic transformation of proteoform and single amino acid variant profiles in colorectal cancer cells after metastasis and discovered potentially critical proteoforms that influence colorectal cancer metastasis. The study employed liquid chromatography fractionation and capillary electrophoresis-tandem mass spectrometry for comprehensive characterization of proteoforms in colorectal cancer cells, improving the number of proteoform identification from human cancer cells by nearly five folds compared to previous studies. Improving our understanding of the underlying molecular mechanisms of colorectal cancer metastasis, the novel technique employed in this study has paved the way for high-throughput and deep top-down proteomics of human cells.
The study is from an exciting collaboration among three research groups, Dr. Liangliang Sun at MSU, Dr. Amanda Hummon at the Ohio State University, and Dr. Xiaowen Liu at Tulane University. The work was funded by the National Cancer Institute (NCI) through grant R01CA247863. The three research groups will work together and continue bettering our understanding of colorectal cancer.
Dr. Sun’s new article in Science Advances: https://www.science.org/doi/10.1126/sciadv.abq6348
For more information on the Human Proteoform Project: https://www.science.org/doi/10.1126/sciadv.abk0734